deepmd_lammps训练和运行镜像
内含ch4简单例子,也可用来训练自己的模型,3080ti版本
 0
00元/小时
v2.0
v3.0
镜像简介
本镜像集成DeepMD机器学习力场与LAMMPS分子动力学模拟环境,内置CH4示例与自定义模型训练支持,针对3080Ti等GPU进行优化,提供开箱即用的配置与脚本。适用于材料计算、分子动力学研究及力场开发等场景,助力科研人员与开发者高效开展仿真模拟与机器学习融合的探索。
镜像使用指南
1、在社区镜像区域,选择镜像:
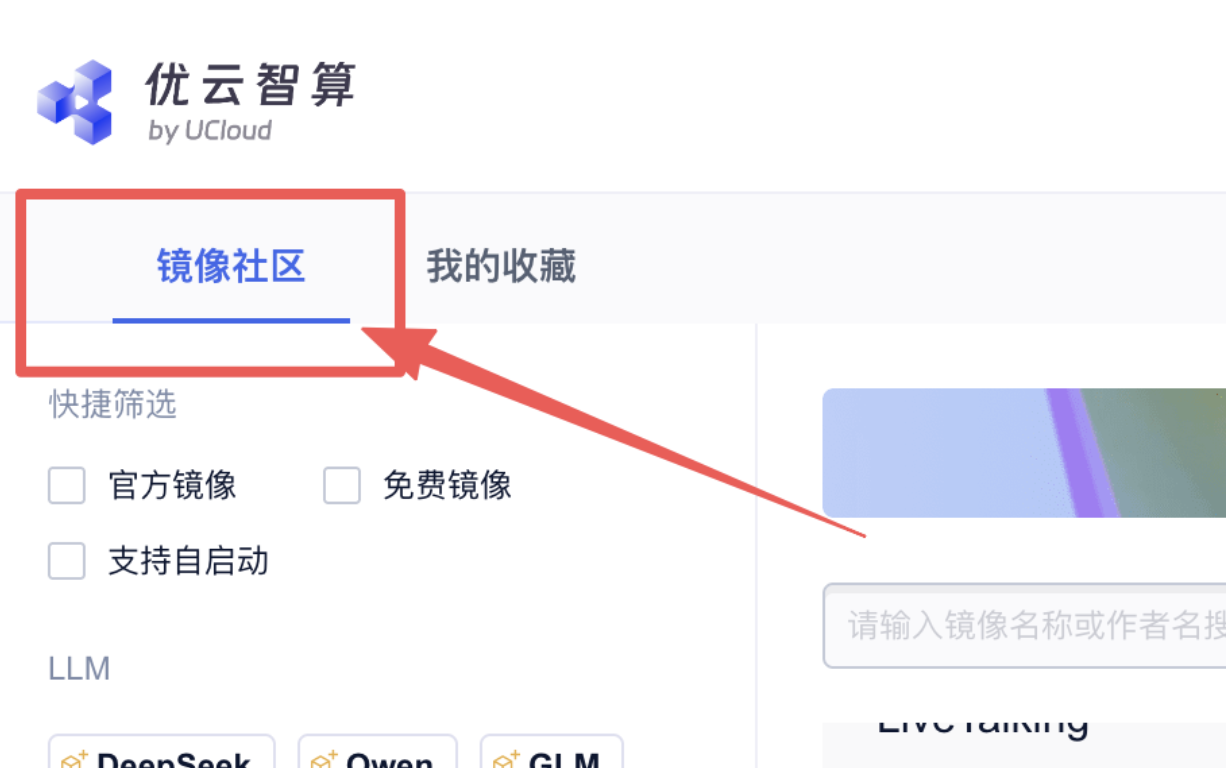
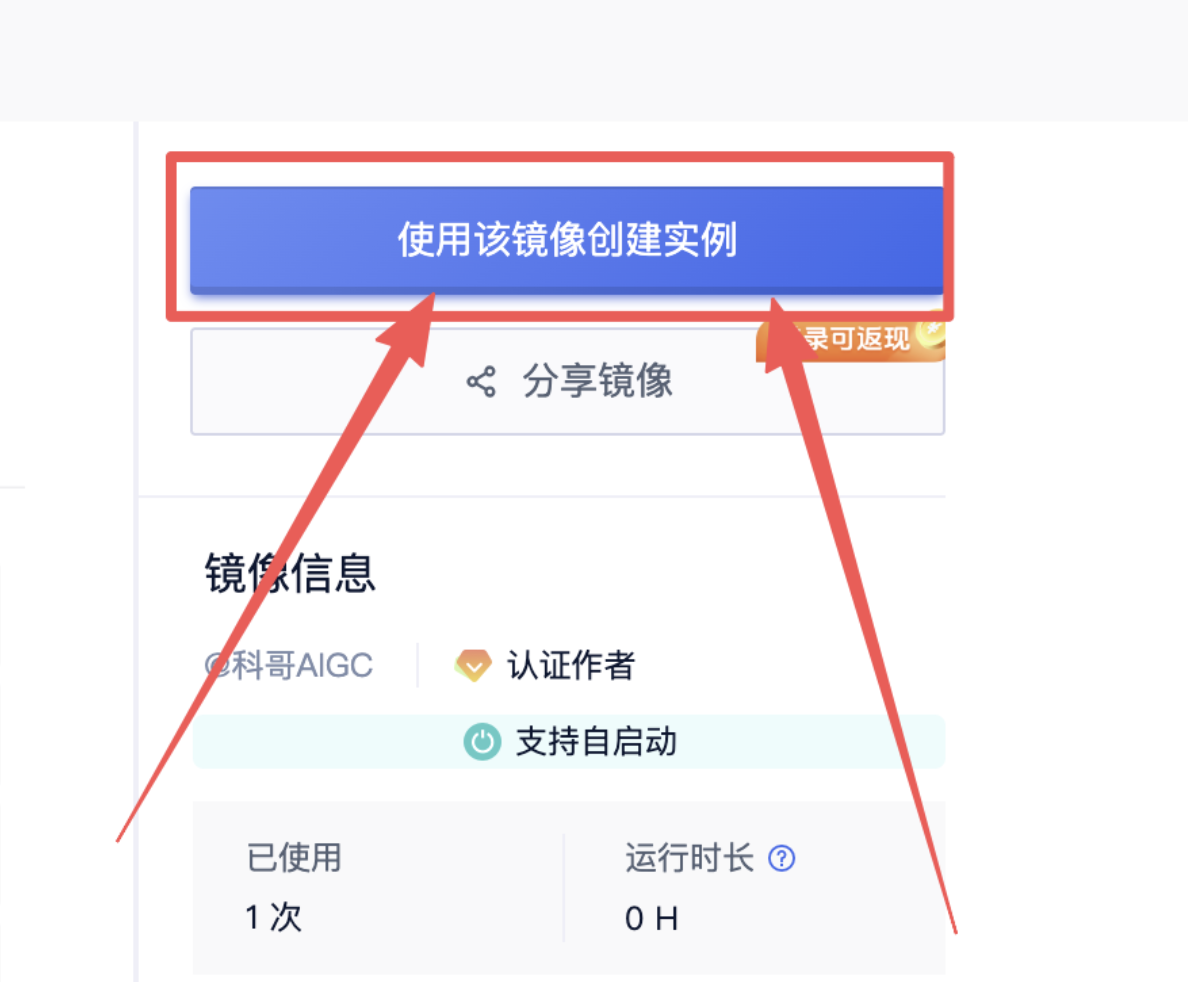
2、在打开的新页面 点击“使用该镜像创建实例”:
2、在打开的新页面 点击“使用该镜像创建实例”:
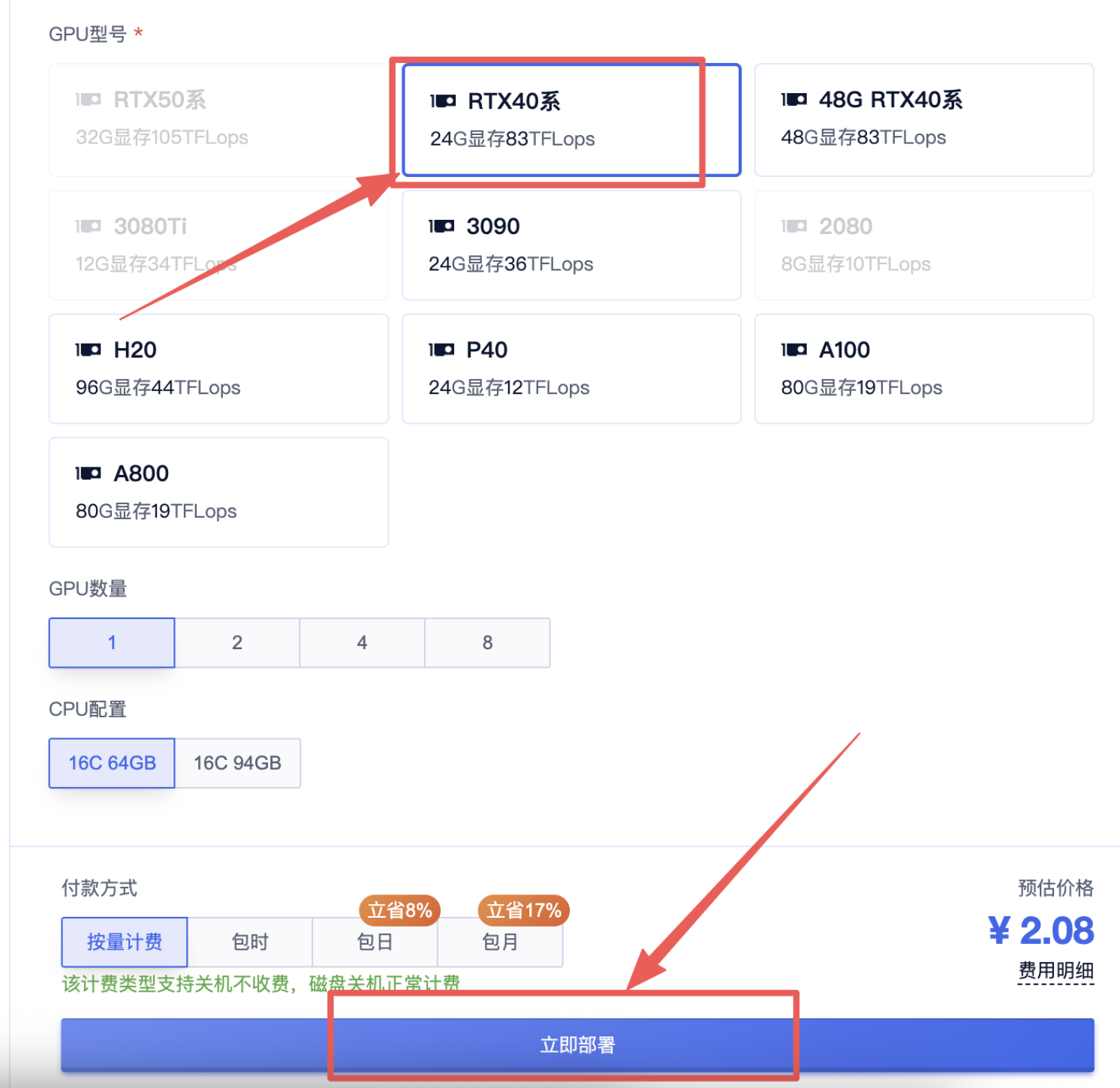
3、选择一个合适的显卡【GPU】根据情况选择,点击“开始部署”,然后等待部署完成:
3、选择一个合适的显卡【GPU】根据情况选择,点击“开始部署”,然后等待部署完成:
 4、 点击【Jupyterlab】
4、 点击【Jupyterlab】
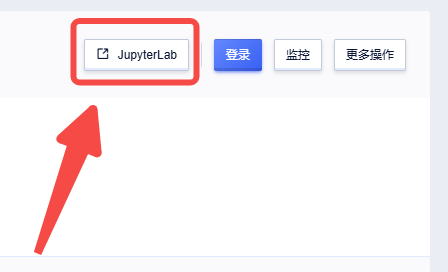
进入工作目录
cd /workspace/test/CH4
$ls
00.data 01.train 02.lmp
进入data目录 cd /workspace/test/CH4/00.data 使用脚本转化文件 python p.py
$python t.py
# the data contains 200 frames
# the training data contains 160 frames
# the validation data contains 40 frames
转化完毕进入训练目录 cd /workspace/test/CH4/01.train 运行deepmd训练 dp --tf train input.json
$dp --pt train input.json
To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, DP_INTRA_OP_PARALLELISM_THREADS, and DP_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
[2025-12-26 11:29:59,692] DEEPMD INFO DeePMD version: 3.1.3.dev21+gb98f6c596.d20251226
[2025-12-26 11:29:59,692] DEEPMD INFO Configuration path: input.json
[2025-12-26 11:29:59,711] DEEPMD INFO _____ _____ __ __ _____ _ _ _
[2025-12-26 11:29:59,711] DEEPMD INFO | __ \ | __ \ | \/ || __ \ | | (_)| |
[2025-12-26 11:29:59,712] DEEPMD INFO | | | | ___ ___ | |__) || \ / || | | | ______ | | __ _ | |_
[2025-12-26 11:29:59,712] DEEPMD INFO | | | | / _ \ / _ \| ___/ | |\/| || | | ||______|| |/ /| || __|
[2025-12-26 11:29:59,712] DEEPMD INFO | |__| || __/| __/| | | | | || |__| | | < | || |_
[2025-12-26 11:29:59,712] DEEPMD INFO |_____/ \___| \___||_| |_| |_||_____/ |_|\_\|_| \__|
[2025-12-26 11:29:59,712] DEEPMD INFO Please read and cite:
[2025-12-26 11:29:59,712] DEEPMD INFO Wang, Zhang, Han and E, Comput.Phys.Comm. 228, 178-184 (2018)
[2025-12-26 11:29:59,712] DEEPMD INFO Zeng et al, J. Chem. Phys., 159, 054801 (2023)
[2025-12-26 11:29:59,712] DEEPMD INFO Zeng et al, J. Chem. Theory Comput., 21, 4375-4385 (2025)
[2025-12-26 11:29:59,712] DEEPMD INFO See https://deepmd.rtfd.io/credits/ for details.
[2025-12-26 11:29:59,712] DEEPMD INFO -------------------------------------------------------------------------------------------------------------------------
[2025-12-26 11:29:59,712] DEEPMD INFO installed to: /usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/deepmd
[2025-12-26 11:29:59,712] DEEPMD INFO source: v3.1.2-21-gb98f6c59-dirty
[2025-12-26 11:29:59,712] DEEPMD INFO source branch: devel
[2025-12-26 11:29:59,712] DEEPMD INFO source commit: b98f6c59
[2025-12-26 11:29:59,712] DEEPMD INFO source commit at: 2025-12-23 08:15:14 +0000
[2025-12-26 11:29:59,712] DEEPMD INFO use float prec: double
[2025-12-26 11:29:59,712] DEEPMD INFO build variant: cuda
[2025-12-26 11:29:59,712] DEEPMD INFO Backend: PyTorch
[2025-12-26 11:29:59,712] DEEPMD INFO PT ver: v2.8.0+cu128-ga1cb3cc05d4
[2025-12-26 11:29:59,712] DEEPMD INFO Enable custom OP: True
[2025-12-26 11:29:59,712] DEEPMD INFO build with PT ver: 2.8.0
[2025-12-26 11:29:59,712] DEEPMD INFO build with PT inc: /usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/torch/include
[2025-12-26 11:29:59,712] DEEPMD INFO /usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/torch/include/torch/csrc/api/include
[2025-12-26 11:29:59,712] DEEPMD INFO build with PT lib: /usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/torch/lib
[2025-12-26 11:29:59,712] DEEPMD INFO running on: 2d27abf23e30
[2025-12-26 11:29:59,712] DEEPMD INFO computing device: cuda:0
[2025-12-26 11:29:59,712] DEEPMD INFO CUDA_VISIBLE_DEVICES: unset
[2025-12-26 11:29:59,712] DEEPMD INFO Count of visible GPUs: 1
[2025-12-26 11:29:59,712] DEEPMD INFO num_intra_threads: 0
[2025-12-26 11:29:59,712] DEEPMD INFO num_inter_threads: 0
[2025-12-26 11:29:59,712] DEEPMD INFO -------------------------------------------------------------------------------------------------------------------------
[2025-12-26 11:29:59,766] DEEPMD INFO Calculate neighbor statistics... (add --skip-neighbor-stat to skip this step)
[2025-12-26 11:30:00,398] DEEPMD INFO Neighbor statistics: training data with minimal neighbor distance: 1.042950
[2025-12-26 11:30:00,399] DEEPMD INFO Neighbor statistics: training data with maximum neighbor size: [4 1] (cutoff radius: 6.000000)
[2025-12-26 11:30:00,402] DEEPMD INFO Constructing DataLoaders from 1 systems
[2025-12-26 11:30:00,409] DEEPMD INFO Constructing DataLoaders from 1 systems
[2025-12-26 11:30:00,435] DEEPMD INFO Packing data for statistics from 1 systems
[2025-12-26 11:30:00,727] DEEPMD INFO RMSE of energy per atom after linear regression is: 0.003203402867550632 in the unit of energy.
[2025-12-26 11:30:00,729] DEEPMD INFO ---Summary of DataSystem: training -----------------------------------------------
[2025-12-26 11:30:00,729] DEEPMD INFO found 1 system(s):
[2025-12-26 11:30:00,729] DEEPMD INFO system natoms bch_sz n_bch prob pbc
[2025-12-26 11:30:00,729] DEEPMD INFO ../00.data/training_data 5 7 22 1.000e+00 T
[2025-12-26 11:30:00,729] DEEPMD INFO --------------------------------------------------------------------------------------
[2025-12-26 11:30:00,729] DEEPMD INFO ---Summary of DataSystem: validation -----------------------------------------------
[2025-12-26 11:30:00,729] DEEPMD INFO found 1 system(s):
[2025-12-26 11:30:00,729] DEEPMD INFO system natoms bch_sz n_bch prob pbc
[2025-12-26 11:30:00,729] DEEPMD INFO ../00.data/validation_data 5 7 5 1.000e+00 T
[2025-12-26 11:30:00,729] DEEPMD INFO --------------------------------------------------------------------------------------
[2025-12-26 11:30:00,730] DEEPMD INFO Start to train 10000 steps.
[2025-12-26 11:30:01,464] DEEPMD INFO batch 1: trn: rmse = 2.45e+01, rmse_e = 2.10e-01, rmse_f = 7.75e-01, lr = 1.00e-03
[2025-12-26 11:30:01,465] DEEPMD INFO batch 1: val: rmse = 1.59e+01, rmse_e = 4.20e-01, rmse_f = 5.02e-01
[2025-12-26 11:30:01,465] DEEPMD INFO batch 1: total wall time = 0.73 s, eta = 2:02:23
[2025-12-26 11:30:05,251] DEEPMD INFO batch 100: trn: rmse = 4.92e+00, rmse_e = 1.15e+00, rmse_f = 1.55e-01, lr = 1.00e-03
[2025-12-26 11:30:05,252] DEEPMD INFO batch 100: val: rmse = 5.14e+00, rmse_e = 1.14e+00, rmse_f = 1.62e-01
[2025-12-26 11:30:05,252] DEEPMD INFO batch 100: total wall time = 3.79 s, eta =
...
...
...
[2025-12-26 11:36:12,576] DEEPMD INFO batch 9900: val: rmse = 3.33e-01, rmse_e = 6.14e-04, rmse_f = 1.27e-01
[2025-12-26 11:36:12,577] DEEPMD INFO batch 9900: total wall time = 3.81 s, eta = 0:00:03
[2025-12-26 11:36:16,286] DEEPMD INFO batch 10000: trn: rmse = 3.91e-01, rmse_e = 4.56e-04, rmse_f = 1.49e-01, lr = 5.92e-06
[2025-12-26 11:36:16,287] DEEPMD INFO batch 10000: val: rmse = 3.31e-01, rmse_e = 7.47e-04, rmse_f = 1.26e-01
[2025-12-26 11:36:16,287] DEEPMD INFO batch 10000: total wall time = 3.71 s, eta = 0:00:00
[2025-12-26 11:36:16,315] DEEPMD INFO Saved model to model.ckpt-10000.pt
[2025-12-26 11:36:16,317] DEEPMD INFO average training time: 0.0375 s/batch (100 batches excluded)
[2025-12-26 11:36:16,317] DEEPMD INFO Trained model has been saved to: model.ckpt
画图 python pl.py
提取模型dp --pt freeze -o graph.pth && dp --pt compress -i graph.pth -o graph-compress.pth
$dp --pt freeze -o graph.pth && dp --pt compress -i graph.pth -o graph-compress.pth
To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, DP_INTRA_OP_PARALLELISM_THREADS, and DP_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
[2025-12-26 11:43:48,763] DEEPMD INFO DeePMD version: 3.1.3.dev21+gb98f6c596.d20251226
[2025-12-26 11:43:49,552] DEEPMD INFO Saved frozen model to graph.pth
To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, DP_INTRA_OP_PARALLELISM_THREADS, and DP_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
[2025-12-26 11:43:55,358] DEEPMD INFO DeePMD version: 3.1.3.dev21+gb98f6c596.d20251226
[2025-12-26 11:43:55,734] DEEPMD INFO training data with lower boundary: [[-1.69396556 -0. -0. -0. ]
[-1.99835585 -0. -0. -0. ]]
[2025-12-26 11:43:55,735] DEEPMD INFO training data with upper boundary: [[1.62150871 2.91929499 2.91929499 2.91929499]
[0.63613754 2.04202009 2.04202009 2.04202009]]
进入运行目录
cd /workspace/test/CH4/02.lmp
直接运行即可
lmp -in in.lammps
$lmp -in in.lammps
DeePMD-kit: Successfully load libcudart.so.12
LAMMPS (29 Aug 2024 - Update 1)
OMP_NUM_THREADS environment is not set. Defaulting to 1 thread. (src/comm.cpp:98)
using 1 OpenMP thread(s) per MPI task
Reading data file ...
triclinic box = (0 0 0) to (10.114259 10.263124 10.216793) with tilt (0.036749877 0.13833062 -0.056322169)
1 by 1 by 1 MPI processor grid
reading atoms ...
5 atoms
read_data CPU = 0.001 seconds
DeePMD-kit WARNING: Environmental variable DP_INTRA_OP_PARALLELISM_THREADS is not set. Tune DP_INTRA_OP_PARALLELISM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
DeePMD-kit WARNING: Environmental variable DP_INTER_OP_PARALLELISM_THREADS is not set. Tune DP_INTER_OP_PARALLELISM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
DeePMD-kit WARNING: Environmental variable OMP_NUM_THREADS is not set. Tune OMP_NUM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
Summary of lammps deepmd module ...
>>> Info of deepmd-kit:
installed to: /root/opt/deepmd/deepmd-kit/source/build/lammps_cpp
source: v3.1.2-21-gb98f6c59-dirty
source branch: devel
source commit: b98f6c59
source commit at: 2025-12-23 08:15:14 +0000
support model ver.: 1.1
build variant: cuda
build with pt lib: torch;torch_library;/usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/torch/lib/libc10.so;/usr/local/cuda-12.8/targets/x86_64-linux/lib/libnvrtc.so;/usr/local/miniconda3/envs/py312/lib/python3.12/site-packages/torch/lib/libc10_cuda.so
set tf intra_op_parallelism_threads: 0
set tf inter_op_parallelism_threads: 0
>>> Info of lammps module:
use deepmd-kit at: /root/opt/deepmd/deepmd-kit/source/build/lammps_cpp
source:
source branch:
source commit:
source commit at:
build with inc:
build with lib:
load model from: graph-compress.pth to gpu 0
DeePMD-kit WARNING: Environmental variable DP_INTRA_OP_PARALLELISM_THREADS is not set. Tune DP_INTRA_OP_PARALLELISM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
DeePMD-kit WARNING: Environmental variable DP_INTER_OP_PARALLELISM_THREADS is not set. Tune DP_INTER_OP_PARALLELISM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
DeePMD-kit WARNING: Environmental variable OMP_NUM_THREADS is not set. Tune OMP_NUM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
>>> Info of model(s):
using 1 model(s): graph-compress.pth
rcut in model: 6
ntypes in model: 2
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Your simulation uses code contributions which should be cited:
- Type Label Framework: https://doi.org/10.1021/acs.jpcb.3c08419
- USER-DEEPMD package:
The log file lists these citations in BibTeX format.
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Generated 0 of 1 mixed pair_coeff terms from geometric mixing rule
Neighbor list info ...
update: every = 10 steps, delay = 0 steps, check = no
max neighbors/atom: 2000, page size: 100000
master list distance cutoff = 7
ghost atom cutoff = 7
binsize = 3.5, bins = 3 3 3
1 neighbor lists, perpetual/occasional/extra = 1 0 0
(1) pair deepmd, perpetual
attributes: full, newton on
pair build: full/bin/atomonly
stencil: full/bin/3d
bin: standard
Setting up Verlet run ...
Unit style : metal
Current step : 0
Time step : 0.001
Per MPI rank memory allocation (min/avg/max) = 2.559 | 2.559 | 2.559 Mbytes
Step PotEng KinEng TotEng Temp Press Volume
0 -23.990493 0.025852029 -23.964641 50 -641.10206 1060.5429
100 -23.991446 0.026485415 -23.96496 51.225022 -644.77032 1060.5429
200 -23.986636 0.021704341 -23.964931 41.978022 -632.30557 1060.5429
300 -23.982735 0.017708755 -23.965026 34.250222 -607.53496 1060.5429
400 -23.985125 0.019381547 -23.965743 37.485542 -603.67587 1060.5429
500 -23.990947 0.024170816 -23.966777 46.748392 -592.71137 1060.5429
600 -23.991475 0.023923502 -23.967552 46.270066 -561.36786 1060.5429
700 -23.98987 0.021328469 -23.968542 41.251055 -530.90052 1060.5429
800 -23.993125 0.022853515 -23.970271 44.200621 -499.23331 1060.5429
900 -23.999956 0.027318557 -23.972637 52.836388 -468.427 1060.5429
1000 -24.000355 0.025433983 -23.974921 49.191464 -422.6516 1060.5429
1100 -23.994028 0.016934155 -23.977094 32.752081 -363.12412 1060.5429
1200 -23.993768 0.014250093 -23.979518 27.56088 -317.82771 1060.5429
1300 -23.998159 0.016289249 -23.981869 31.504779 -271.77165 1060.5429
1400 -24.000464 0.016833702 -23.98363 32.557796 -216.555 1060.5429
1500 -23.998292 0.013576771 -23.984716 26.258618 -160.10552 1060.5429
1600 -23.997844 0.01237625 -23.985468 23.93671 -103.33492 1060.5429
1700 -24.002667 0.016639614 -23.986027 32.182414 -59.594476 1060.5429
1800 -24.00679 0.020782213 -23.986008 40.194549 -13.053831 1060.5429
1900 -24.004126 0.019065643 -23.98506 36.874559 44.137193 1060.5429
2000 -23.999986 0.016667069 -23.983319 32.235515 96.682798 1060.5429
2100 -23.999324 0.018515979 -23.980808 35.811462 151.4457 1060.5429
2200 -24.001711 0.024465753 -23.977245 47.318825 208.6335 1060.5429
2300 -23.99773 0.025483146 -23.972247 49.286549 272.39989 1060.5429
2400 -23.989875 0.023436371 -23.966438 45.327914 346.21517 1060.5429
2500 -23.989638 0.028280584 -23.961357 54.69703 391.08389 1060.5429
2600 -23.996416 0.038747414 -23.957669 74.940761 431.7077 1060.5429
2700 -23.989793 0.034953093 -23.95484 67.602223 475.97062 1060.5429
2800 -23.980215 0.027243689 -23.952971 52.691588 505.86292 1060.5429
2900 -23.982488 0.029642563 -23.952846 57.331212 542.42658 1060.5429
3000 -23.989982 0.035867898 -23.954114 69.371534 551.55237 1060.5429
3100 -23.990312 0.034532858 -23.955779 66.789454 566.5232 1060.5429
3200 -23.989399 0.031660154 -23.957739 61.233402 595.71727 1060.5429
3300 -23.989988 0.029498076 -23.96049 57.051763 598.37095 1060.5429
3400 -23.988142 0.024589899 -23.963552 47.558934 601.35892 1060.5429
3500 -23.984638 0.018356932 -23.966281 35.503851 607.71301 1060.5429
3600 -23.985172 0.016560721 -23.968612 32.029829 603.11253 1060.5429
3700 -23.990131 0.019098118 -23.971033 36.937368 602.7738 1060.5429
3800 -23.994648 0.021293316 -23.973354 41.183065 587.3801 1060.5429
3900 -23.998062 0.022961842 -23.9751 44.410134 574.89552 1060.5429
4000 -23.998875 0.022635323 -23.97624 43.77862 576.28081 1060.5429
4100 -23.999515 0.022254024 -23.977261 43.041155 557.57159 1060.5429
4200 -23.996989 0.018800744 -23.978188 36.362221 546.84963 1060.5429
4300 -23.995901 0.017069942 -23.978831 33.014704 523.26497 1060.5429
4400 -23.995123 0.016162623 -23.97896 31.259874 499.9238 1060.5429
4500 -23.995133 0.016313376 -23.978819 31.551442 481.74798 1060.5429
4600 -23.995432 0.017132848 -23.978299 33.136369 449.62586 1060.5429
4700 -23.996223 0.019324574 -23.976899 37.375353 423.98681 1060.5429
4800 -23.997193 0.02296525 -23.974228 44.416726 405.11768 1060.5429
4900 -23.996856 0.026390678 -23.970465 51.041793 379.62463 1060.5429
5000 -23.993684 0.027621606 -23.966062 53.42251 367.77872 1060.5429
Loop time of 44.3104 on 1 procs for 5000 steps with 5 atoms
Performance: 9.749 ns/day, 2.462 hours/ns, 112.840 timesteps/s, 564.201 atom-step/s
98.9% CPU use with 1 MPI tasks x 1 OpenMP threads
MPI task timing breakdown:
Section | min time | avg time | max time |%varavg| %total
---------------------------------------------------------------
Pair | 44.264 | 44.264 | 44.264 | 0.0 | 99.89
Neigh | 0.0061518 | 0.0061518 | 0.0061518 | 0.0 | 0.01
Comm | 0.012446 | 0.012446 | 0.012446 | 0.0 | 0.03
Output | 0.0071103 | 0.0071103 | 0.0071103 | 0.0 | 0.02
Modify | 0.01554 | 0.01554 | 0.01554 | 0.0 | 0.04
Other | | 0.005606 | | | 0.01
Nlocal: 5 ave 5 max 5 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Nghost: 130 ave 130 max 130 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Neighs: 0 ave 0 max 0 min
Histogram: 1 0 0 0 0 0 0 0 0 0
FullNghs: 20 ave 20 max 20 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Total # of neighbors = 20
Ave neighs/atom = 4
Neighbor list builds = 500
Dangerous builds not checked
Total wall time: 0:00:47
@tty

镜像信息
已使用9 次
运行时长
33 H
镜像大小
40GB
最后更新时间
2026-01-26
支持卡型
RTX40系3080Ti3090
+3
框架版本
CUDA版本
12.8
应用
JupyterLab: 8888
版本
v2.0
2026-01-26
v3.0
2026-01-26
